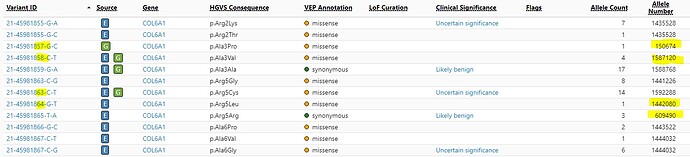
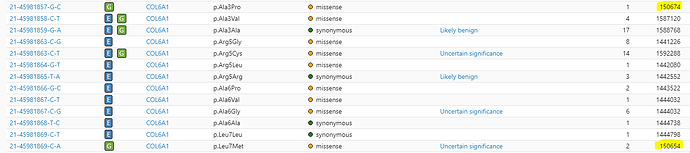
Why is there such a huge difference in total allele numbers between neighboring bases?
Hi, I was wondering if someone from the gnomad team could answer this.
Thanks! ![]()
Hello,
Apologies for the delay and thank you for your patience. The key to the explanation to your question ought to be under ‘Source’ column ; looking at the first pair, 21-…857-G-C is only called in Genomes (‘G’ in Source) while 21-…858-C-T is called in both Exomes and Genomes (‘E’ & ‘G’ under Source). As such, the -G-C variant includes the AN only from Genomes. (In general: even if a variant’s site is covered in Exomes or Genomes coverage, if AC=0 then AN is not included, since the variant is not called). The -C-T variant nextdoor then includes the AN from both Exomes and Genomes, since it is called by both.
Let me know if this answers your question, and let me know if you have any further comments or questions or concerns.
Dear Daniel,
Thank you for the explanation.
I still do not understand why no Exomes or Genomes are included if AC=0. I mean, the reported allele frequencies are then skewed and wrong if thousands of cases that do not contain the alternative allele are not included in the calculation.
Hello again,
I understand your complaint about this as far as data design and display goes. You are not the first person to reach out to us about this (though none have used the browser).
There is a technical explanation: to expand upon the previous message, we do not joint call our exome and genome data. We have separate tables for variants called in exome and variants called in genomes, which were done separately and from different sets of individuals. They can be downloaded at our downloads page. In those tables, for your example 21-45981857-G-C | Source: Genomes | p.Ala3Pro , it is present in the genomes table but does not exist in the exome variants table, as there were no non-reference calls at this site, meaning that variant information about that site for that variant (such as AN) was not emitted. Due to this, it was misleading to say its AC=0 in exomes, when more accurately it has no AC in exomes or has an AC=None.
Let me know if you have any further questions about this or any comments or questions or concerns or any notes for the team.
Hi,
Why do the positions I reported in December still seem to have the wrong total allele count number in gnomAD v4.1.0?
I thought the issue discussed above would be solved by the new version.
Hi, I just came across the same bug in my own work and reported it to the browser team. I came to the forum to see if anyone else reported this issue and found your post. This bug is isolated to the browser, the hail tables are updated. We figured out the source of the bug and it should be updated in the next couple days.